From Bioblast
The MitoPedia terminology is developed continuously in the spirit of Gentle Science.
| Term | Abbreviation | Description |
|---|---|---|
| AMPK | AMPK | AMP-activated protein kinase is a regulatory protein which acts as crucial cellular energy sensor by sensing AMP, ADP and/or Ca2+ levels in response to metabolic stresses or drug administration. |
| ATP synthase | CV | ATP synthase or F-ATPase (F1FO-ATPase; the use of Complex V is discouraged) catalyzes the endergonic phosphorylation of ADP to ATP in an over-all exergonic process that is driven by proton translocation along the protonmotive force. The ATP synthase can be inhibited by oligomycin. |
| ATPases | ATPases are enzymes that hydrolyse ATP, releasing ADP and inorganic phosphate. The contamination of isolated mitochondria with ATPases from other organelles and endogenous adenylates can lead to the production of ADP, which can stimulate respiration. This situation would lead to an overestimation of LEAK respiration measured in the absence of ADP, L(n) and subsequent inhibition of respiration by oligomycin, L(Omy). | |
| Aconitase | Aco | Aconitase is a TCA cycle enzyme that catalyzes the reversible isomerization of citrate to isocitrate. Also, an isoform is also present in the cytosol acting as a trans-regulatory factor that controls iron homeostasis at a post-transcriptional level. |
| Acyl-CoA dehydrogenase | ACAD | Acyl-CoA dehydrogenases ACADs are localized in the mitochondrial matrix. Several ACADs are distinguished: short-chain (SCAD), medium-chain (MCAD), and long-chain (LCAD). ACAD9 is expressed in human brain. ACADs catalyze the reaction
|
| Acyl-CoA oxidase | Acyl-CoA oxidase is considered as a rate-limiting step in peroxysomal β-oxidation, which carries out few β-oxidation cycles, thus shortening very-long-chain fatty acids (>C20). Electrons are directly transferred from FADH2 to O2 with the formation of H2O2. | |
| Adenine nucleotide translocase | ANT | The adenine nucleotide translocator, ANT, exchanges ADP for ATP in an electrogenic antiport across the inner mt-membrane. The ANT is inhibited by atractyloside, carboxyatractyloside and bongkrekik acid. The ANT is a component of the phosphorylation system. |
| Adenylate kinase | ADK | Adenylate kinase, which is also called myokinase, is a phosphotransferase enzyme that is located in the mitochondrial intermembrane space and catalyzes the rephosphorylation of AMP to ADP in the reaction ATP + AMP ↔ ADP + ADP. |
| Alternative oxidase | AOX | Alternative quinol oxidases AOX are membrane-bound enzymes capable of supporting cyanide- and antimycin A-resistant mitochondrial respiration. AOX catalyzes the oxidation of ubiquinol and the reduction of oxygen to water in a four-electron process. As this bypasses several proton-translocating steps, induction of this alternative pathway is associated with a reduction of ATP production per oxygen consumed. AOX is found in most plants (including microalgae), many fungi and protists, but is not expressed in animals. AOX is inhibited by salicylhydroxamic acid (SHAM). Expression and activity of the enzyme are modified by environmental conditions such as temperature, oxidative stress, nutrient availability, and pathogens such as viruses. |
| Carnitine O-octanoyltransferase | COT | Carnitine O-octanoyltransferase is a mitochondrial enzyme that transfers carnitine to octanoyl-CoA to form Coenzyme A and octanoylcarnitine: Octanoyl-CoA + L-carnitine ↔ CoA + L-octanoylcarnitine. |
| Carnitine acetyltransferase | CrAT | Carnitine acetyltransferase (CrAT) is located in the mitochondrial matrix and catalyses the formation of acetyl-carnitine from acetyl-CoA and L-carnitine and thus regulates the acetyl-CoA/free CoA ratio which is essential for pyruvate dehydrogenase complex (PDC) activity. |
| Carnitine acyltransferase | Carnitine acyltransferases mediate the transport of long-chain fatty acids across the inner mt-membrane by binding them to carnitine. First, long-chain fatty acids are activated by an energy-requiring step in which the fatty acid ester of CoA is formed enzymatically at the expense of ATP. The fatty acids then pass through the inner mt-membrane and enter the mitochondria as carnitine esters (acylcarnitines). The fatty acyl group is then transferred from carnitine to intramitochondrial CoA and the resulting fatty acyl CoA is used as a substrate in the fatty acid oxidation (FAO) cycle in the mt-matrix. | |
| Carnitine palmitoyltransferase I | CPT-I | Carnitine palmitoyltransferase I (CPT-I, also known as carnitine acyltransferase I) is a regulatory enzyme in mitochondrial long-chain acyl-CoA uptake and further oxidation. CPT-I is associated with the mt-outer membrane mtOM and catalyses the formation of acylcarnitines from acyl-CoA and L-carnitine. In the next step, acyl-carnitines are transported to the mitochondrial matrix via carnitine-acylcarnitine translocase in exchange for free carnitine. In the inner side of the mtIM carnitine palmitoyltransferase II converts the acyl-carnitines to carnitine and acyl-CoAs. There are three enzyme isoforms: CPT-1A (liver type), CPT-1B (muscle type), CPT-1C (brain type). Isoforms have significantly different kinetic and regulatory properties. Malonyl-CoA is an endogenous inhibitor of CPT-I. |
| Carnitine palmitoyltransferase II | CPT-II | Carnitine palmitoyltransferase II (CPT-II, also known as carnitine acyltransferase II) is part of the carnitine shuttle which is responsible for the mitochondrial transport of long-chain fatty acids. CPT-II is located on the inner side of the mtIM and converts the acylcarnitines (produced in the reaction catalyzed by carnitine palmitoyltransferase I) to carnitine and acyl-CoAs, which undergo ß-oxidation in the mitochondrial matrix. Free carnitines are transported out of the mitochondrial matrix in exchange for acyl-carnitines via an integral mtIM protein carnitine-acylcarnitine translocase (CACT). Short- and medium-chain fatty acids do not require the carnitine shuttle for mitochondrial transport. |
| Carnitine-acylcarnitine translocase | CACT | Carnitine-acylcarnitine translocase (CACT) is part of the carnitine shuttle which mediates the mitochondrial transport of long-chain fatty acids where the fatty acid oxidation occurs. CACT is an internal mt-IM protein and transports acylcarnitines into the mitochondrial matrix in exchange for free carnitine. |
| Catalase | Ctl | Catalase catalyzes the dismutation of hydrogen peroxide to water and oxygen. Perhaps all cells have catalase, but mitochondria of most cells lack catalase. Cardiac mitochondria are exceptional in having mt-catalase activity (rat heart mitochondria: Radi et al 1991; mouse heart mitochondria: Rindler et al 2013). Hydroxylamine is an inhibitor of catalase, which is also inhibited by cyanide and azide. Mitochondrial respiration medium MiR05 was developed considering the intracellular conditions of mitochondria in living cells. In mitochondrial preparations, enzymes and substrates present in the cytosol (such as catalase) are diluted when the plasma membrane is removed. Therefore, the addition of catalase is recommended when working with mitochondrial preparations, to consume any H2O2 generated during the assay. |
| Catalytic activity | kat | Catalytic activity of an enzyme is measured by an enzyme assay and is expressed in units of katal (kat [mol∙s-1]). More commonly (but not conforming to SI units or IUPAC recommendations) enzyme activity is expressed in units U [mol∙min-1]. |
| Choline dehydrogenase | Choline dehydrogenase (EC 1.1.99.1) is bound to the inner mt-membrane, oxidizes choline in kidney and liver mitochondria, with electron transfer into the Q-junction, and is thus part of the Electron transfer pathway. Analogous to succinate dehydrogenase (CII), electron transfer from choline dehydrogenase is FAD-linked downstream to Q. Choline is an ET-pathway substrate types 3. | |
| Citrate synthase | CS | Condensation of oxaloacetate with acetyl-CoA yields citrate as an entry into the TCA cycle. CS is located in the mt-matrix. CS activity is frequently used as a functional marker of the amount of mitochondria (mitochondrial elementary marker, mtE) for normalization of respiratory flux. |
| Coenzyme | A coenzyme or cosubstrate is a cofactor that is attached loosely and transiently to an enzyme, in contrast to a prosthetic group that is attached permanently and tightly. The coenzyme is required by the corresponding enzyme for its activity (IUPAC definition). A coenzyme is 'a low-molecular-weight, non-protein organic compound participating in enzymatic reactions as dissociable acceptor or donor of chemical groups or electrons' (IUPAC definition). | |
| Coenzyme A | CoA | Coenzyme A is a coenzyme playing an essential role in the tricarboxylic acid cycle (oxidation of pyruvate to acetyl-CoA) and fatty acid oxidation. CoA is a thiol that reacts with carboxylic acids to form CoA-activated thioesters. |
| Cofactor | A cofactor is 'an organic molecule or ion (usually a metal ion) that is required by an enzyme for its activity. It may be attached either loosely (coenzyme) or tightly (prosthetic group)' (IUPAC definition). | |
| Complex I | CI | Complex I, NADH:ubiquinone oxidoreductase (EC 1.6.5.3), is an enzyme complex of the Electron transfer pathway, a proton pump across the inner mt-membrane, responsible for electron transfer to ubiquinone from NADH formed in the mt-matrix. CI forms a supercomplex with Complex III. There is a widespread ambiguity on the 'lonely H+ (the lonely hydron)' surrounding Complex I: CI ambiguities. |
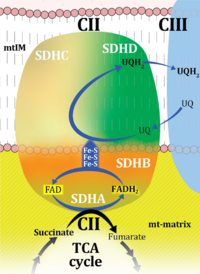
| Complex II | CII | Complex II or succinate:quinone oxidoreductase (SQR) is the only membrane-bound enzyme in the TCA cycle and is part of the electron transfer pathway. The reversible oxidoreduction of succinate and fumarate is catalyzed in a soluble domain and coupled to the reversible oxidoreduction of quinol and quinone in the mitochondrial inner membrane. CII consists in most species of four subunits. The flavoprotein succinate dehydrogenase is the largest polypeptide of CII, located on the matrix face of the mt-inner membrane. Succinate:quinone oxidoreductases (SQRs, SDHABCD) favour oxidation of succinate and reduction of quinone in the canonical forward direction of the TCA cycle and electron transfer into the Q-junction. In contrast, quinol:fumarate reductases (QFRs, fumarate reductases, FRDABCD) tend to operate in the reverse direction reducing fumarate and oxidizing quinol. |
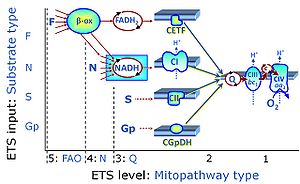
| Complex II ambiguities | CII ambiguities | The current narrative that the reduced coenzymes NADH and FADH2 feed electrons from the tricarboxylic acid (TCA) cycle into the mitochondrial electron transfer system can create ambiguities around respiratory Complex CII. Succinate dehydrogenase or CII reduces FAD to FADH2 in the canonical forward TCA cycle. However, some graphical representations of the membrane-bound electron transfer system (ETS) depict CII as the site of oxidation of FADH2. This leads to the false believe that FADH2 generated by electron transferring flavoprotein (CETF) in fatty acid oxidation and mitochondrial glycerophosphate dehydrogenase (CGpDH) feeds electrons into the ETS through CII. In reality, NADH and succinate produced in the TCA cycle are the substrates of Complexes CI and CII, respectively, and the reduced flavin groups FMNH2 and FADH2 are downstream products of CI and CII, respectively, carrying electrons from CI and CII into the Q-junction. Similarly, CETF and CGpDH feed electrons into the Q-junction but not through CII. The ambiguities surrounding Complex II in the literature call for quality control, to secure scientific standards in current communications on bioenergetics and support adequate clinical applications. |
| Complex III | CIII | Complex III or coenzyme Q : cytochrome c - oxidoreductase, sometimes also called the cytochrome bc1 complex is a complex of the electron transfer pathway. It catalyzes the reduction of cytochrome c by oxidation of coenzyme Q (CoQ) and the concomitant pumping of 4 protons from the cathodic (negative) mitochondrial matrix to the anodic (positive) intermembrane space. |
| Complex IV | CIV | Complex IV or cytochrome c oxidase is the terminal oxidase of the mitochondrial electron transfer system, reducing oxygen to water, with reduced cytochrome c as a substrate. Concomitantly to that, CIV pumps protons against the electrochemical protonmotive force. CIV is frequently abbreviated as COX or CcO. It is the 'ferment' (Atmungsferment) of Otto Warburg, shown to be related to the cytochromes discovered by David Keilin. |
| Creatine kinase | CK | The mitochondrial creatine kinase, also known as phosphocreatine kinase (CPK), facilitates energy transport with creatine and phosphocreatine as diffusible intermediates. |
| Dicarboxylate carrier | DIC | The dicarboxylate carrier is a transporter which catalyses the electroneutral exchange of malate2- (or succinate2-) for inorganic phosphate, HPO42-. |
| Dihydro-orotate dehydrogenase | DhoDH | Dihydro-orotate dehydrogenase is an electron transfer complex of the inner mitochondrial membrane, converting dihydro-orotate (Dho) into orotate, and linking electron transfer through the Q-junction to pyrimidine synthesis and thus to the control of biogenesis. |
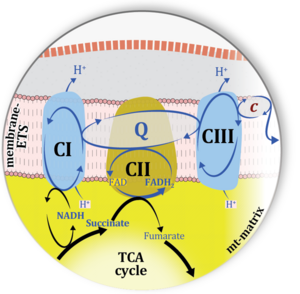
| Electron transfer pathway | ET pathway | In the mitochondrial electron transfer pathway (ET pathway) electrons are transferred from externally supplied reduced fuel substrates to oxygen. Based on this experimentally oriented definition (see ET capacity), the ET pathway consists of (1) the membrane-bound ET pathway with respiratory complexes located in the inner mt-membrane, (2) TCA cycle and other mt-matrix dehydrogenases generating NADH and succinate, and (3) the carriers involved in metabolite transport across the mt-membranes. » MiPNet article |
| Electron-transferring flavoprotein Complex | CETF | Electron-transferring flavoprotein Complex (CETF) is a respiratory Complex localized at the matrix face of the inner mitochondrial membrane, supplies electrons to Q, and is thus an enzyme Complex of the mitochondrial Electron transfer pathway (ET-pathway). CETF links the ß-oxidation cycle with the membrane-bound electron transfer system in fatty acid oxidation (FAO). |
| Fumarase | FH | Fumarase or fumarate hydratase (FH) is an enzyme of the tricarboxylic acid cycle catalyzing the equilibrium reaction between fumarate and malate. Fumarase is found not only in mitochondria, but also in the cytoplasm of all eukaryotes. |
| Glutamate dehydrogenase | mtGDH | Glutamate dehydrogenase, located in the mitochondrial matrix (mtGDH), is an enzyme that converts glutamate to α-ketoglutarate [1]. mtGDH is not part of the TCA cycle, but is involved in glutaminolysis as an anaplerotic reaction. |
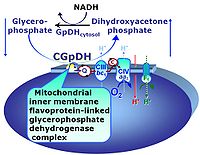
| Glycerophosphate dehydrogenase Complex | CGpDH | Glycerophosphate dehydrogenase complex (CGpDH) is a Complex of the electron transfer-pathway localized at the outer face of the mt-inner membrane. CGpDH is thus distinguished from cytosolic GpDH. CGpDH oxidizes glycerophosphate to dihydroxyacetone phosphate and feeds two electrons into the Q-junction, thus linked to an ET pathway level 3 control state. |
| Glycerophosphate shuttle | Gp shuttle | The glycerophosphate shuttle makes cytoplasmic NADH available for mitochondrial oxidative phosphorylation. Cytoplasmic NADH reacts with dihydroxyacetone phosphate catalyzed by cytoplasmic glycerophosphate dehydrogenase. On the outer face of the inner mitochondrial membrane, glycerophosphate dehydrogenase complex (mitochondrial glycerophosphate dehydrogenase) oxidizes glycerophosphate back to dihydroxyacetone phosphate, a reaction not generating NADH but reducing a flavin prosthesic group. The reduced flavoprotein transfers its reducing equivalents into the Q-junction, thus representing a ET pathway level 3 control state. |
| Hexokinase | HK | The hexokinase catalyzes the phosphorylation of D-glucose at position 6 by ATP to yield D-glucose 6-phosphate as well as the phosphorylation of many other hexoses like D-fructose, D-mannose, D-glucosamine. |
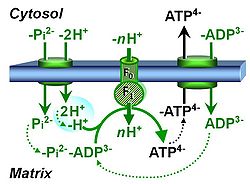
| Hydrogen ion pump | Mitochondrial hydrogen ion pumps — frequently referred to as "proton pumps" — are large enzyme complexes (CI, CIII, CIV, ATP synthase) spanning the mt-inner membrane mtIM, partially encoded by mtDNA. CI, CIII and CIV are H+ pumps that drive hydrogen ions against the electrochemical protonmotive force pmF and thus generating the pmF, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CV) is a H+ pump that utilizes the exergy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
| Isocitrate dehydrogenase | IDH | Isocitrate dehydrogenase forms 2-oxoglutarate from isocitrate in the TCA cycle. |
| Kynurenine hydroxylase | Kynurenine hydroxylase (kynurenine 3-monooxygenase) is located in the outer mitochondrial membrane. Kynurenine hydroxylase catalyzes the chemical reaction: L-kynurenine + NADPH + H+ + O2 ↔ 3-hydroxy-L-kynurenine + NADP+ + H2O Kynurenine hydroxylase belongs to the family of oxidoreductases acting on paired donors, with O2 as oxidant and incorporation or reduction of oxygen. The oxygen incorporated need not be derived from O2 with NADH or NADPH as one donor, and incorporation of one atom of oxygen into the other donor. This enzyme participates in tryptophan metabolism. It employs one cofactor, FAD. | |
| Lactate dehydrogenase | LDH | Lactate dehydrogenase is a glycolytic marker enzyme in the cytosol, regenerating NAD+ from NADH and pyruvate, forming lactate. |
| Malate dehydrogenase | mtMDH | Mitochondrial malate dehydrogenase is localized in the mitochondrial matrix and oxidizes malate, generated from fumarate by fumarase, to oxaloacetate, reducing NAD+ to NADH+H+ in the TCA cycle. Malate is added as a substrate in most N-pathway control states. |
| Malate transport | Carriers for malate: | |
| Malate-aspartate shuttle | The malate-aspartate shuttle involves the glutamate-aspartate carrier and the 2-oxoglutarate carrier exchanging malate2- for 2-oxoglutarate2-. Cytosolic and mitochondrial malate dehydrogenase and transaminase complete the shuttle for the transport of cytosolic NADH into the mitochondrial matrix. It is most important in heart, liver and kidney. | |
| Malic enzyme | mtME | Malic enzyme (ME; EC 1.1.1.40) catalyzes the oxidative decarboxylation of L-malate to pyruvate with the concomitant reduction of the dinucleotide cofactor NAD+ or NADP+ and a requirement for divalent cations (Mg2+ or Mn2+) as cofactors.
NAD(P)+ + L-malate2- <--> NAD(P)H + pyruvate- + CO2 Three groups of ME are distinguished (i) NAD+- and (ii) NADP+-dependent ME specific for NAD+ or NADP+, respectively, and (iii) NAD(P)+- dependent ME with dual specificity for NAD+ or NADP+ as cofactor. Three isoforms of ME have been identified in mammals: cytosolic NADP+-dependent ME (cNADP-ME or ME1), mitochondrial NAD(P)+-dependent ME (mtNAD-ME or ME2; with NAD+ or NADP+ as cofactor, preference for NAD+ under physiological conditions), and mitochondrial NADP+-dependent ME (mtNADP-ME or ME3). mtNAD-ME plays an important role in anaplerosis when glucose is limiting, particularly in heart and skeletal muscle. Tartronic acid (hydroxymalonic acid) is an inhibitor of ME. |
| Malonyl-CoA synthase | Malonyl-CoA synthase or ACSF3 protein is a mitochondrial fatty-acyl-CoA synthase found in mammals. Traditionally, malonyl-CoA is formed from acetyl-CoA by the action of acetyl-CoA carboxylase. However, Witkowski et al (2011) showed that mammals express malonyl-CoA Synthase (ACSF3) with enzymatic activity in the presence of malonate (Complex II inhibitor) and methylmalonate. | |
| Matrix-ETS | matrix-ETS | The component of the electron transfer system located in the mitochondrial matrix (matrix-ETS) is distringuished from the ETS bound to the mt-inner membrane (membrane-ETS). Electron transfer and corresponding OXPHOS capacities are classically studied in mitochondrial preparations as oxygen consumption supported by various fuel substrates undergoing partial oxidation in the mt-matrix, such as pyruvate, malate, succinate, and others. |
| Membrane-bound ET pathway | mET-pathway | The membrane-bound electron transfer pathway (mET pathway) consists in mitochondria mainly of respiratory complexes CI, CII, electron transferring flavoprotein complex (CETF), glycerophosphate dehydrogenase complex (CGpDH), and choline dehydrogenase, with convergent electron flow at the Q-junction (Coenzyme Q), and the two downstream respiratory complexes connected by cytochrome c, CIII and CIV, with oxygen as the final electron acceptor. The mET-pathway is the terminal (downstream) module of the mitochondrial ET pathway and can be isolated from the ET-pathway in submitochondrial particles (SmtP). |
| Mitochondrial ATP-sensitive K+ channel | mtKATP | The mitochondrial ATP-sensitive K+ channel (mtKATP or mitoKATP). |
| Mitochondrial marker | mt-marker | Mitochondrial markers are structural or functional properties that are specific for mitochondria. A structural mt-marker is the area of the inner mt-membrane or mt-volume determined stereologically, which has its limitations due to different states of swelling. If mt-area is determined by electron microscopy, the statistical challenge has to be met to convert area into a volume. When fluorescent dyes are used as mt-marker, distinction is necessary between mt-membrane potential dependent and independent dyes. mtDNA or cardiolipin content may be considered as a mt-marker. Mitochondrial marker enzymes may be determined as molecular (amount of protein) or functional properties (enzyme activities). Respiratory capacity in a defined respiratory state of a mt-preparation can be considered as a functional mt-marker, in which case respiration in other respiratory states is expressed as flux control ratios. » MiPNet article |
| Mitochondrial marker enzymes | Mitochondrial marker enzymes are enzymes that are specifically present in mitochondria, in the mt-matrix, the inner mt-membrane, the inter-membrane space, or the outer mt-membrane. | |
| Monoamine oxidase | MAO | Monoamine oxidases are enzymes bound to the outer membrane of mitochondria and they catalyze the oxidative deamination of monoamines. Oxygen is used to remove an amine group from a molecule, resulting in the corresponding aldehyde and ammonia. Monoamine oxidases contain the covalently bound cofactor FAD and are, thus, classified as flavoproteins. |
| Nagarse | Nagarse is a broad specifity protease from bacteria used to promote breakdown of the cellular structure of "hard" tissues such as skeletal muscle or heart mucsle that cannot be homogenized easily without treatment with a protease. Nagarse is frequently used in protocols for isolating mitochondria from muscle tissue. | |
| Nitric oxide synthase | NOS | Nitric oxide synthase, NOS, catalyzes the production of nitric oxide (NO•), which is a reactive nitrogen species. There are four types of NOS: neuronal NOS (nNOS), endothelial NOS (eNOS), inducible NOS (iNOS) and mitochondrial NOS (mtNOS). |
| Oxoglutarate dehydrogenase | OgDH | Oxoglutarate dehydrogenase (α-ketoglutarate dehydrogenase) is a highly regulated enzyme of the tricarboxylic acid cycle. It catalyses the conversion of oxoglutarate (alpha-ketoglutarate) to succinyl-CoA, reduces NAD+ to NADH and thus links to Complex I in the Electron transfer-pathway. OgDH is activated by low Ca2+ (<20 µM) but inactivated by high Ca2+ (>100 µM). OgDH is an important source of ROS. |
| Phosphate carrier | PiC | The phosphate carrier (PiC) is a proton/phosphate symporter which transports negatively charged inorganic phosphate across the inner mt-membrane. The transport can be described either as symport of H+ with Pi, or antiport of hydroxide anion against Pi. The phosphate carrier is a component of the phosphorylation system. |
| Phosphoenolpyruvate carboxykinase | PEPCK | Phosphoenolpyruvate carboxykinase (PEPCK) catalyzes the anabolic reaction of oxaloacetate (Oxa) to phosphoenolpyruvate at the expense of GTP. PEPCK is a cytoplasmatic enzyme involved in gluconeogenesis in mouse and rat liver, but 'is found in the mitochondria in the rabbit and chicken, and in both cytoplasm and mitochondria in the guinea pig' (Lehninger 1970). In many anoxia-resistant animals, PEPCK plays an important catabolic role under severe hypoxia and anoxia at the PEPCK branchpoint (Hochachka, Somero 2002), feeding malate into the reversed TCA cycle: malate is dismutated to pyruvate catalyzed by malic enzyme in the oxidative direction, and to fumarate in the reductive direction, leading to formation of succinate and ATP under anoxia (Gnaiger 1977). |
| Phosphorylation pathway | DT | The phosphorylation pathway (phosphorylation system) is the functional unit utilizing the protonmotive force to phosphorylate ADP (D) to ATP (T), and may be defined more specifically as the P»-system. The P»-system consists of adenine nucleotide translocase, phosphate carrier, and ATP synthase. Mitochondrial adenylate kinase, mt-creatine kinase and mt-hexokinase constitute extended components of the P»-system, controlling local AMP and ADP concentrations and forming metabolic channels. Since substrate-level phosphorylation is involved in the TCA-cycle, the P»-system includes succinyl-CoA ligase (GDP to GTP or ADP to ATP). |
| Proline dehydrogenase | ProDH | Proline dehydrogenase (ProDH), L-proline:quinone oxidoreductase, is located on the inner side of the mtIM, oxidizing proline to delta-1-pyrroline-5-carboxylate, with reduction of FAD to FADH2 and direct entry into the Q-junction, exerting an additive effect of convergent pathways. ProDH is widely distributed in a variety of organisms, is a source of ROS, and may play a role in carcinogenesis. |
| Prosthetic group | A prosthetic group is a cofactor that is attached permanently and tightly or even covalently to an enzyme and that is regenerated in each enzymatic turnover. Thus a prostethic group is distinguished from a coenzyme or cosubstrate that is attached loosely and transiently. Like a coenzyme, the prosthetic group is required by an enzyme for its activity. A prosthetic group is 'a tightly bound, specific nonpolypeptide unit in a protein determining and involved in its biological activity' (IUPAC definition). FMN/FMNH2 and FAD/FADH2 are prosthetic groups of Complex I and Complex II, respectively. | |
| Proton pump | Mitochondrial proton pumps are large enzyme complexes (CI, CIII, CIV, CV) spanning the inner mt-membrane, partially encoded by mtDNA. CI, CIII and CIV are proton pumps that drive protons against the electrochemical protonmotive force, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CIV) is a proton pump that utilizes the energy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
| Pyruvate carboxylase | PC | Pyruvate carboxylase synthesizes oxaloacetate from pyruvate and CO2 as an anaplerotic reaction in the mitochondrial matrix of the liver and kidney of higher animals, representing an alternative to the malic enzyme pathway to oxaloacetate or the phosphoenolpyruvate carboxykinase reaction (compare glyoxylate cycle in plants and microorganisms). Carboxylation of pyruvate to oxaloacetate requires Mg-ATP. Acetyl CoA is a strong positive modulator. PC can form pyruvate from oxaloacetate to remove an excess of oxaloacetate which inhibits succinate dehydrogenase. |
| Pyruvate carrier | The monocarboxylic acid pyruvate- is exchanged electroneutrally for OH- by the pyruvate carrier. H+/anion symport is equivalent to OH-/anion antiport. | |
| Pyruvate dehydrogenase | PDH | Pyruvate dehydrogenase is the first component enzyme of the pyruvate dehydrogenase complex, which catalyzes oxidative decarboxylation of pyruvate in the mt-matrix, and yields acetyl-CoA. PDH is known as the mitochondrial gatekeeper in the core energy pathway of electron flow into the tricarboxylic acid cycle. |
| Pyruvate dehydrogenase complex | PDHC | Oxidative decarboxylation of pyruvate is catalyzed by the pyruvate dehydrogenase complex in the mt-matrix, and yields acetyl-CoA. |
| P»-system | P»system | The ADP-ATP phosphorylation system or P»-system. See Phosphorylation system. |
| Q-junction | The Q-junction is a junction for convergent electron flow in the Electron transfer pathway (ET-pathway) from type N substrates and mt-matrix dehydrogenases through Complex I (CI), from type F substrates and FA oxidation through electron-transferring flavoprotein Complex (CETF), from succinate (S) through Complex II (CII), from glycerophosphate (Gp) through glycerophosphate dehydrogenase Complex (CGpDH), from choline through choline dehydrogenase, from dihydro-orotate through dihydro-orotate dehydrogenase, and other enzyme Complexes into the Q-cycle (ubiquinol/ubiquinone), and further downstream to Complex III (CIII) and Complex IV (CIV). The concept of the Q-junction, with the N-junction and F-junction upstream, provides the rationale for defining Electron-transfer-pathway states and categories of SUIT protocols. | |
| Respiratory complexes | CI, CII, CIII, CIV, CETF, CGpDH, .. | Respiratory Complexes are membrane-bound enzymes consisting of several subunits which are involved in energy transduction of the respiratory system. » MiPNet article |
| Reverse electron flow from CII to CI | RET | Reverse electron flow from CII to CI stimulates production of ROS when mitochondria are incubated with succinate without rotenone in the LEAK state at a high mt-membrane potential. Depolarisation of the mt-membrane potential (e.g. after ADP addition to stimulate OXPHOS) leads to inhibition of RET and therefore, decrease of RET-initiated ROS production. RET can be also measured when mitochondria are respiring using Gp without rotenone in the LEAK state. Addition of IQ-side inhibitors (ubiquinone-binding side of CI) of CI usually block RET. The following SUIT protocols allow you to measure RET-initiated H2O2 flux in mitochondrial preparations: SUIT-009 and SUIT-026. |
| Sirtuins | Sirt | Sirtuins are NAD+-dependent deacetylases which play a prominent role as metabolic regulators. Their dependence on intracellular levels of NAD+ (NAD+ activates sirtuin activity, whereas NADH inhibits it) makes them suitable as sensors that can detect cellular energy status. » MiPNet article |
| Succinate dehydrogenase | SDH | Succinate dehydrogenase is a TCA cycle enzyme converting succinate to fumarate while reducing FAD to FADH2. SDH is the largest component of the mt-inner membrane Complex II (CII) and thus part of the TCA cycle and electron transfer pathway. |
| Succinate transport | The dicarboxylate carrier catalyses the electroneutral exchange of succinate2- for HPO4-2-. | |
| Succinyl-CoA ligase | SUCLA, SUCLG | Succinyl-CoA ligase (SUCLA or SUCLG) is a TCA cycle enzyme converting succinyl-CoA + ADP or (GDP) + Pi to succinate + ATP (GTP). Two different isoforms exsist: SUCLA (EC: 6.2.1.5) is the ATP-forming isoenzyme, SUCLG (EC: 6.2.1.4) is the GTP-forming isoenzyme. Both reactions are reversible. This reaction is attributed to mitochondrial substrate-level phosphorylation, which is considered as an alternative way of ATP synthesis because it is partially independent from the respiratory chain and from the mitochondrial proton motive force. |
| Sulfide quinone reductase | SQR | Sulfide quinone reductase (SQR) is involved in electron transfer from sulfide which is used as a hydrogen donor by the mitochondrial respiratory system. SQR is associated with a dioxygenase and a sulfur transferase to release thiosulfate (H2S2O3). |
| Sulfite oxidase | SO | Sulfite oxidase (SO) is a dimeric enzyme, located in the intermembrane space of mitochondria, with each monomer containing a single Mo cofactor and cyt b5-type heme [1]. SO catalyzes the oxidation of sulfite to sulfate as the terminal step in the metabolism of sulfur amino acids and is vital for human health. Inherited mutations in SO result in severe neurological problems, stunted brain growth, and early death [2].
Function: SO catalyzes the terminal reaction in the oxidative degradation of sulfur amino acids with the formation of a sulfate, electrons pass to cytochrom c and are further utilized in the respiratory system. Sulfite + O2 + H2O --> Sulfate + H2O2 Localization: The level of expression of SO differs in various tissues with main predominant localization in liver, kidney, skeletal muscle, heart, placenta, and brain in humans and liver, kidney, heart, brain, and lung in rats [3]. Deficiency: SO is vital for metabolic pathways of sulfur amino acids (cysteine and methionine). Complete lack of this enzyme, typically caused by gene mutation, leads to lethal disease called sulfite oxidase deficiency characterized by neurological abnormalities with brain atrophy. |
| Superoxide dismutase | SOD | Mammalian superoxide dismutase (SOD) exists in three forms, of which the Mn-SOD occurs in mitochondria (mtSOD, SOD2; 93 kD homotetramer) and many bacteria, in contrast to the Cu-Zn forms of SOD (cytosolic SOD1, extracellular SOD3 anchored to the extracellular matrix and cell surface). Superoxide anion (O2•-) is a major reactive oxygen species (ROS) which is dismutated by SOD to oxygen and H2O2. |
| Thioredoxin reductase | TrxR | Thioredoxin reductase (TrxR) is a family of enzymes able to reduce thioredoxin in mammals. |
| Tricarboxylate carrier | The tricarboxylate carrier in the inner mt-membrane exchanges malate2- for citrate3- or isocitrate3-, with co-transport of H+. |