Difference between revisions of "Categories of SUIT protocols"
| Line 2: | Line 2: | ||
|abbr=SUIT-catg | |abbr=SUIT-catg | ||
|description=[[File:SUIT-catg_MitoPathway types.jpg|right|200px]] | |description=[[File:SUIT-catg_MitoPathway types.jpg|right|200px]] | ||
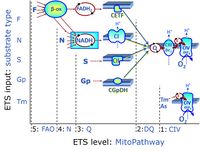
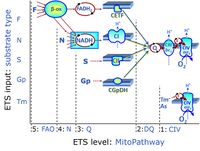
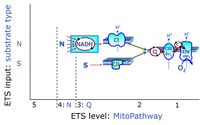
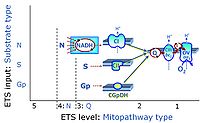
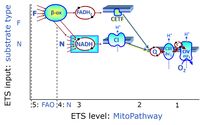
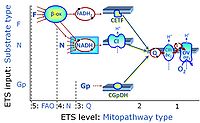
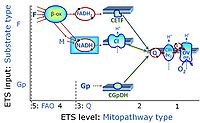
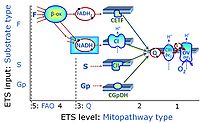
'''Categories of SUIT protocols''' group SUIT protocols according to all | '''Categories of SUIT protocols''' group SUIT protocols according to all substrate types involved in a protocol (F, N, S, Gp), independent of the sequence of titrations of substrates and inhibitors which define the [[pathway control state]]s. ROX states may or may not be included in a SUIT protocol, which does not change its category. Similarly, the [[CIV]] assay may or may not be added towards the end of a SUIT protocol, without effect on the category of a SUIT protocol. | ||
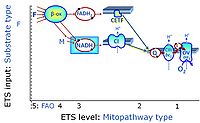
* '''F''' - ETS-level 5: [[FADH2 |FADH<sub>2</sub>]]-linked substrates (FAO) with obligatory support by the N-linked pathway. | * '''F''' - ETS-level 5: [[FADH2 |FADH<sub>2</sub>]]-linked substrates (FAO) with obligatory support by the N-linked pathway. | ||
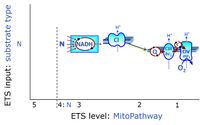
* '''N''' - ETS-level 4: [[NADH]]-linked substrates (CI-linked). | * '''N''' - ETS-level 4: [[NADH]]-linked substrates (CI-linked). | ||
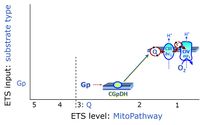
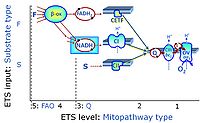
* '''S''' - ETS-level 3: [[Succinate]] (CII-linked). | * '''S''' - ETS-level 3: [[Succinate]] (CII-linked). | ||
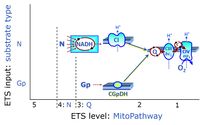
* '''Gp''' - ETS-level 3: [[Glycerophosphate]] (CGpDH-linked). | * '''Gp''' - ETS-level 3: [[Glycerophosphate]] (CGpDH-linked). | ||
» [[#Categorization of SUIT protocols: ETS pathway control states |'''MiPNet article''']] | » [[#Categorization of SUIT protocols: ETS pathway control states |'''MiPNet article''']] | ||
| Line 16: | Line 16: | ||
}} | }} | ||
__TOC__ | __TOC__ | ||
= | = Categories of SUIT protocols and ETS pathway control states = | ||
{{Publication | {{Publication | ||
|title=Gnaiger E ( | |title=Gnaiger E (2017) Categories of SUIT protocols and ETS pathway control states. MiPNet 2016-03-20, edited 2016-08-21, 2017-03-11. | ||
|info=[[MiPNet21.06 SUIT RP]] | |info=[[MiPNet21.06 SUIT RP]] | ||
|authors=OROBOROS | |authors=OROBOROS | ||
|year= | |year=2017 | ||
|journal=MiPNet | |journal=MiPNet | ||
|abstract=There are many ways to define groups of SUIT protocols. The complexity of SUIT protocols is primarily determined by the large number of | |abstract=There are many ways to define groups of SUIT protocols. The complexity of SUIT protocols is primarily determined by the large number of [[pathway control state]]s and fuel substrates involved, compared to only three well defined [[coupling control state]]s. Whereas the [[SUIT protocol names]] specify all sequential steps in a SUIT protocol, the categories of SUIT protocols reduce this diversity to a list of substrate types involved. | ||
|mipnetlab=AT Innsbruck OROBOROS | |mipnetlab=AT Innsbruck OROBOROS | ||
}} | }} | ||
| Line 34: | Line 34: | ||
== Towards a library of SUIT protocols == | == Towards a library of SUIT protocols == | ||
:::: At the present stage of development of the [[MitoPedia:_SUIT#Library_of_SUIT_protocols |Library of SUIT protocols]] as part of the [[MitoFit Quality Control System]], five ETS pathway types are considered. | :::: At the present stage of development of the [[MitoPedia:_SUIT#Library_of_SUIT_protocols |Library of SUIT protocols]] as part of the [[MitoFit Quality Control System]], the substrate types linked to five ETS pathway types are considered. | ||
::::* '''F-junction''' on the pathway level of converging FADH<sub>2</sub>- and NADH-linked dehydrogenases, including beta-oxidationthe and segments of the TCA cycle. | ::::* '''F-junction''' on the pathway level of converging FADH<sub>2</sub>- and NADH-linked dehydrogenases, including beta-oxidationthe and segments of the TCA cycle. | ||
::::* '''N-junction''' on the pathway level of NADH-linked dehydrogenases, including the TCA cycle. | ::::* '''N-junction''' on the pathway level of NADH-linked dehydrogenases, including the TCA cycle. | ||
| Line 41: | Line 41: | ||
== | == Single pathway type == | ||
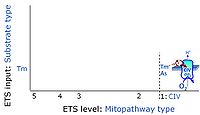
=== ETS-level 1: CIV === | === ETS-level 1: CIV === | ||
| Line 59: | Line 59: | ||
== | == Multiple ETS pathways with FNSGp == | ||
[[File: SUIT-catg_FNSGpCIV.jpg|200px]] | [[File: SUIT-catg_FNSGpCIV.jpg|200px]] | ||
== | == Multiple ETS pathways with NS == | ||
=== NS === | === NS === | ||
| Line 78: | Line 78: | ||
== | == Multiple ETS pathways with N (without S) == | ||
=== FN === | === FN === | ||
| Line 90: | Line 90: | ||
== | == Multiple ETS pathways without N == | ||
:::: Addition of malate alone (M without P or G) is not considered as substrate type N. However, high mt-malic enzyme activity requires a change of this concept, when M alone represents an ETS ccompetent substrate state. Low concentration of M may be used to support FAO, whereas a higher concentration of M may be required for N-junction respiratory capacity to override FAO capacity; this needs corresponding kinetic analyses ([[SUIT test protocols]]). | :::: Addition of malate alone (M without P or G) is not considered as substrate type N. However, high mt-malic enzyme activity requires a change of this concept, when M alone represents an ETS ccompetent substrate state. Low concentration of M may be used to support FAO, whereas a higher concentration of M may be required for N-junction respiratory capacity to override FAO capacity; this needs corresponding kinetic analyses ([[SUIT test protocols]]). | ||
Revision as of 12:27, 11 March 2017
Description
Categories of SUIT protocols group SUIT protocols according to all substrate types involved in a protocol (F, N, S, Gp), independent of the sequence of titrations of substrates and inhibitors which define the pathway control states. ROX states may or may not be included in a SUIT protocol, which does not change its category. Similarly, the CIV assay may or may not be added towards the end of a SUIT protocol, without effect on the category of a SUIT protocol.
- F - ETS-level 5: FADH2-linked substrates (FAO) with obligatory support by the N-linked pathway.
- N - ETS-level 4: NADH-linked substrates (CI-linked).
- S - ETS-level 3: Succinate (CII-linked).
- Gp - ETS-level 3: Glycerophosphate (CGpDH-linked).
Abbreviation: SUIT-catg
Reference: MiPNet21.06 SUIT RP
MitoPedia concepts:
MiP concept,
SUIT concept
Categories of SUIT protocols and ETS pathway control states
| Gnaiger E (2017) Categories of SUIT protocols and ETS pathway control states. MiPNet 2016-03-20, edited 2016-08-21, 2017-03-11. |
Abstract: There are many ways to define groups of SUIT protocols. The complexity of SUIT protocols is primarily determined by the large number of pathway control states and fuel substrates involved, compared to only three well defined coupling control states. Whereas the SUIT protocol names specify all sequential steps in a SUIT protocol, the categories of SUIT protocols reduce this diversity to a list of substrate types involved.
• O2k-Network Lab: AT Innsbruck OROBOROS
Labels: MiParea: Instruments;methods
Pathway: F, N, S, Gp, CIV, NS, Other combinations, ROX
HRR: Theory
Towards a library of SUIT protocols
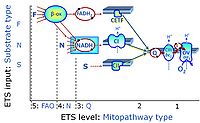
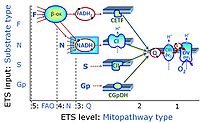
- At the present stage of development of the Library of SUIT protocols as part of the MitoFit Quality Control System, the substrate types linked to five ETS pathway types are considered.
- F-junction on the pathway level of converging FADH2- and NADH-linked dehydrogenases, including beta-oxidationthe and segments of the TCA cycle.
- N-junction on the pathway level of NADH-linked dehydrogenases, including the TCA cycle.
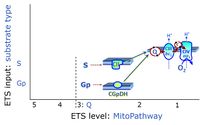
- Q-junction on the pathway level of electron transfer complexes converging at the Q-junction (S and Gp).
- Tm on the single step level of cytochrome c oxidase (CIV), the terminal step in the aerobic electron transfer system. Tm can be included or excluded at the end of a SUIT protocol. To simplify the categorization, Tm is not considered in this system of SUIT protocols.
- At the present stage of development of the Library of SUIT protocols as part of the MitoFit Quality Control System, the substrate types linked to five ETS pathway types are considered.
Single pathway type
ETS-level 1: CIV
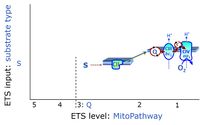
ETS-level 3: S, CII-pathway to Q
ETS-level 3: Gp, CGpDH-pathway to Q
ETS-level 4: N, N-junction or CI-pathway to Q
ETS-level 5: F, FAO, F-junction, CETF- supported by CI-pathway to Q
Multiple ETS pathways with FNSGp
Multiple ETS pathways with NS
NS
FNS
NSGp
FNSGp
Multiple ETS pathways with N (without S)
FN
NGp
FNGp
Multiple ETS pathways without N
- Addition of malate alone (M without P or G) is not considered as substrate type N. However, high mt-malic enzyme activity requires a change of this concept, when M alone represents an ETS ccompetent substrate state. Low concentration of M may be used to support FAO, whereas a higher concentration of M may be required for N-junction respiratory capacity to override FAO capacity; this needs corresponding kinetic analyses (SUIT test protocols).